Mammalian Speciation
Natura non agit saltatim. Gottfried Wilhelm Leibniz, 1698.
Natura non facit saltum . Charles Darwin, 1859.
Discontinuity in evolution . Francis Galton, 1894.
Evolution by macro-mutations . De Vries, 1901.
Punctuated equilibria . N. Eldredge and S.J. Gould, 1972.
Introduction
The mammalian placenta has many different phenotypes – macroscopic and microscopic. Grosser (1927) made a beginning with the use of placental forms as a means to fathom evolutionary mammalian relationships. Since then, numerous efforts have been made ( e.g. Mossman, 1987) to employ the placental form and, especially, the “barrier” between the maternal and fetal bloods for a better understanding of evolution amongst mammalian species. Most recently, Carter and Enders (2004) put a new effort into such a classification, whilst also employing the new genetic insights gained by molecular biologists.
A further exploration of the use of the placenta for evolutionary relationships, however, is not the purpose of this brief essay; we have done so once in the past with respect to primate placentas (Benirschke & Miller, 1982). Moreover, many of these features can be gleaned from exploring the pages in this web site. Here, though, I want to put into ‘print' these pages so as to discuss my view of mammalian speciation , particularly the ascendancy of primates and, quite especially, that of the human evolution from chimpanzee ancestors. One reason is the following:
In lectures I am often asked: “ Why is it that so many animals are becoming extinct but no new ones are being created? Aren't extinction of animals and evolution of species parts of the same process? ”
This is indeed the case, but the process of speciation, i.e. the evolution of a new mammalian species, has not yet been witnessed and much speculation surrounds this in a shroud of mystery, and the processes that lead to the evolution of new species remain the causes of endless speculations. To be sure, some new species are being discovered , as for instance new lemur species in Madagascar, a new peccary in Brazil and so on; but those animals have been there for a long time, they are not ‘new species' in the sense here discussed, they merely have only now been found for the first time. So, what then are the possible reasons for this situation? What processes are currently truly known that ‘create' a new mammalian species?
Ever since the publication of Darwin 's book (1859) – THE BOOK - this topic has aroused more questions than it has received answers. The earlier of the two recent books written by Michael Ruse (2003) has weighed the pros and cons that have followed the publication of ‘THE BOOK'. In this volume he summarized the views of the adherents of Darwin 's view and he referred to the gradualistic construct of Darwin 's hypothesis for evolution. In addition, the anti-Darwinian thoughts are carefully reviewed within the context of historical time. This book is truly worth a good read, as the evolution of the Darwin hypothesis and the numerous critics of it are so beautifully laid out.
One often hears that the ‘adaptation to changing environment' can be witnessed. A case in point for instance would be the acquisition of antibiotic resistance of bacteria. But the numerous examples of this kind are not truly speciation events; those are relatively minor genetic changes of an organism and they are usually without consequences for reproduction. More massive ones can be witnessed by comparing the great variety of dog phenotypes, features that were fixed through specific inbreeding efforts. The Staphylococcus aureus that becomes resistant to penicillin therapy is still a staphylococcus, not a new ‘species' and so are the dogs only ‘races', not new species. (Not to mention that several definitions of “species” are only poorly if at all applicable to bacteria where horizontal exchange of genes is rampant even between very distantly related organisms).
Of course, the denomination of ‘species' and even of ‘genera' by us humans has a major impact on understanding these issues. In his searching review on Hybrids, van Gelder (1977) has laid this out quite explicitly. Indeed, is ‘hybridizability' even a good criterion for the separation of species? If it were, such a requirement would surely have to include the continued fertility of some of the resultant hybrids. (Please see Barton, 2001). Furthermore, numerous similar genetic changes as are demonstrated by the dog races have been witnessed in many taxa, and others are still being described. Weiner (2005) produced an admirable summary of such events, ranging from the peppered moth in England to new studies on Galapagos finches, sticklebacks, salmon and other taxa. The topic of Weiner's essay was to ‘ watch evolution in action'. Yes, it considered the Darwinian mode of gradual selection and adaptation to a changing environment etc., one that elicits changes (adaptations) in the species under study. In some cases (finches), the genes that allow the change to proceed have even become known. The question of how to define a species has been much debated, but most recently, Baker & Bradley (2006) have strongly argued for the “Genetic Species Concept” and reviewed most of the past history of various other concepts of what constitutes a species. They defined as ‘genetic species ….a group of genetically compatible interbreeding natural populations that is genetically isolated from other such groups’.
But most of these examples are not events that create a new species when examined more closely. At least in my view those are not processes of speciation. Speaking in broader terms then, Evolution is not necessarily synonymous with Speciation . The question really is whether adaptation to a changing environment can truly create new species or whether we must invoke different mechanisms for their creation? Such a question possibly embodies the notion of ‘saltation' or punctuation, a concept that Darwin (1895) had denied. His proposed mechanism of evolution was a more gradual and adaptive one. It contrasted with the idea of saltation. Darwin 's early critic and his so-called ‘bulldog', T.H. Huxley, already proclaimed saltation as being a probable event in the speciation of animals. But the concept was later much further refined and it was then proclaimed as gospel by Eldredge & Gould (1972). They created the notion of the ‘Punctuated Equilibrium' (with its component of long periods of ‘Stasis'). Tattersall (2002) has further discussed the implications that are implied in that concept for pre- homo species.
There are many ways by which one can envisage saltational speciation to occur, an important one of which is through significant chromosomal structural change. Major chromosomal structural changes, with their implicit founder effects, would subsequently have to become adaptive, essentially so by the future acquisition of beneficial (and also of some neutral) mutations. But this process of mutations is complex and has been most competently addressed with entirely new concepts by Kirschner & Gerhart (2005). These authors argue that major ‘conserved core processes' (mitochondria, cytoskeleton, cell adhesion, etc.) arise only rarely but, once in place, they serve for the occurrence of ‘facilitated variation', a concept that they have espoused in this book. This is an interesting and most persuasive concept that well may explain the “Plausibility of Life', as they contend in their fascinating book.
It was M.J.D. White (1978) who estimated that over 90% of speciation events are accompanied by significant karyotypic changes and that these have played a primary role in the speciation process! Moreover, he reasoned that few genetic changes occur during the speciation event, rather, that most of those would have occurred after the speciation had been completed. Ernst Mayr, grandmaster of “Species”, considered that allopatric speciation was the most important mechanism and was skeptical of the stasipatric speciation concept as proposed by White. It is said, however, that he changed his mind to some extent when confronted with the evolution of cichlids in the Great Lakes of Africa. These fish are numerous (+/- 1,500 species), have apparently speciated from small ancestries and occupy very special niches in their new environments. Numerous studies have addressed the mechanisms that might have led to their rapid and perhaps increasingly frequent speciation events over the past. Temperature, transposable elements, sex-related mutations and color changes, locations of NORs, ploidy changes have all been implicated and additional studies are ongoing (Lande et al., 2001; Seehausen et al., 1997, Brinn et al., 2004; Genner & Turner, 2005). Despite often identical chromosome numbers, some cichlid species do not hybridize fertilely, while others do. It will be of interest to learn in the future what the precise mechanisms of speciation are in cichlids, and importantly, what their chromosomal structures are. Many species possess 22 or 24 chromosomes but much higher numbers are also reported (Chiarelli & Capanna, 1973).
It may also be relevant here that Gould (2002) in his massive tome discusses the ‘bushiness' of the human ancestral tree and likens it to that of the horse. He draws (incorrectly, I believe) the conclusion that only one species of horse now exists (overlooking the fact that onager, the zebras, kiang, and donkey most likely derived from the horse ancestors and while not ‘horses', they are nevertheless equids); he then likened this to the bushiness of human evolution, and consequently to Homo sapiens, of which only one species now exists. From the former ‘bushiness' of evolving Homo species, he suggested that as recently as 40,000 years ago three forms (species) were concurrent : Homo neanderthalensis, Homo erectus , and Homo sapiens , of which only the latter survives (pp. 909-912). [I might parenthetically mention here that if indeed chromosomal change is an underlying and principal mechanism of speciation, then it seems to me most improbable that further human speciation (in the sense of the word species used here) is possible because of the extraordinary vagility of humans and the massive crossbreeding that now occurs in Homo sapiens ]. Gould then goes on to vigorously deny the possibility of a ‘multiregional' origin of H. sapiens and, on p. 916, he says:” The change from Homo erectus did not occur in a gradual and global flux throughout the range of Homo erectus , but as an event of speciation, geologically rapid under punctuated equilibrium and local in geography (probably in Africa).”
-----------------------------------------------
The impetus to write this personal view of chromosomal evolution in mammals actually comes from my reading the searching new books by the philosopher Michael Ruse (2003, 2005). In these books, Ruse explores the societal evolution of beliefs regarding ‘ Darwinism ' as opposed to those of ‘ Intelligent Design' or ‘ Creation ' and places these ideas into their appropriate time frames as well as the social climates during the last two centuries.
I will state here at the outset that I view as the one major mechanism of mammalian speciation (and only one but the most important one) a significant and fundamental change of chromosome number/structure and, among these especially, the processes of chromosomal fusion. In most species this is accomplished by a Robertsonian fusion of acrocentric elements. Chromosomal translocations and possibly other major chromosomal rearrangements may play an additional part in some instances. King (1993) has eloquently stated this in his comprehensive book on Speciation by saying:
“ Numerous recently published books or conference proceedings have either downgraded the significance of chromosomal speciation or simply ignored it…. to think that much of the data presented in this book did not exist ”.
Later on he wrote: “ The concept of the founder population is most important to the possibility of chromosomal speciation…. Not only demonstrates it the primacy of chromosome change in establishing divergence between species, it also shows the significance of these differences in establishing profound reproductive isolation .”
The first concept, Robertsonian fusion, implies that two acrocentric chromosomes fuse in an individual animal, causing it to have a reduced chromosome number with the result that one new metacentric element has been formed. This fusion probably would have caused temporarily a degree of reduced fertility. Nevertheless, if it did occur in a genetically relatively ‘disease-free' individual, its offspring would, through the laws of meiotic chromosomal segregation, ultimately produce some individuals that are homozygous for the translocation chromosome. The bottleneck that has thus arisen will succeed only when no major (lethal) genetic abnormalities exist in the animal's genome. It is thus probably a priori an uncommon event. Eventually then, these new homozygous individuals will have an even, but reduced number of chromosomes. This will thereby create the necessary barrier to continued (at least extensive) gene flow with the ancestral stock. Numerous examples exist for this method of speciation to have occurred in the past, only a few of which are listed here for some mammalian species. King (1993) has summarized most of these processes in an eminently readable and well-documented text Moreover, the recent essay by Coyne and Orr (2004) further supports this notion as being a major process of evolution.
Whether chromosomal inversions also form a significant barrier to gene flow will depend on the location of the breakpoints in chromosomes that are involved and on other factors; they are discussed further below only with the example of orang utans in which one major pericentric inversion must have occurred that now separates Sumatran from Bornean orang utans. This is the pericentric inversion of chromosome #2 that has not led to hybrid sterility. In the chimpanzee, a reciprocal translocation must have occurred with chromosomes # 12, 13, the one that has led to the formation of human chromosome # 2; an event that is further discussed below.
These mechanisms, of courses do not preclude that many other non-chromosomal structural events can also participate in the mammalian speciation process, a notion that is also discussed in some detail by King (1993) but is there considered within different contexts. For instance, tigers and lions are surely different ‘species' by our current nomenclature, but they have essentially similar chromosome sets (2n=38) and they also hybridize. This is also the case for the Dromedary and the Bactrian camel (2n=74). It even includes the South American camelidae (2n=74). Interestingly, dromedary and Bactrian camel also ably hybridize fertilely, as do the South American camelidae, while gene flow may have essentially stopped, largely for geographic reasons.
These considerations jibe with the thoughts of van Gelder (1977) who believes that we have made artificial ‘species' denominations when ‘races' would have been sufficient. The Sumatran and Bornean orang utans hybridize fertilely, despite the fact that they have a pericentric chromosomal inversion of chromosome #2 that distinguishes these ‘species' or more properly races (Ryder & Chemnick, 1993). Indeed, these former subspecies have recently been assigned species ranks ( Groves , 2001). The Bornean orang remains as Pongo pygmaeus while the Sumatran orang becomes Pongo abelii. Even some of the numerical rearrangements, as in the Przewalski's horse (2n=66) compared with that of the domestic horses (2n=64), do not preclude fertility of the hybrids produced (see Pennisi, 2001). This is presumably so because of the minimal structural differences of banding patterns that now exists in the involved chromosomes, the linearity of genes, which is probably an essential aspect for chromosomal pairing at meiosis.
The more recent study of diverse genomes by modern and sophisticated techniques, however, shows that chromosomal speciation is very much more complex. Eichler & Sankoff (2003) have provided a survey of this complexity. They highlighted transposable elements, extensive duplications, break points for rearrangements, etc. for genomic evolution. They will become relevant when more detailed genome analysis of related species is accomplished and they can thus not be further considered here. My main point of departure is to ask: “Why is it that all human karyotypes display exactly the same chromosomal fusion event, two ancestral chromosomes having joined and now being represented by our new chromosomes # 2, chromosomes that are still separate in the chimpanzee?” Is it not logical that it must have arisen in a single ancestral individual chimpanzee (yielding 2n=47) whose subsequent ‘inbreeding' then led to our karyotype of 46 chromosomes, a reduction from 48 from the chimpanzee? How else can one possibly envisage that all humans have essentially the same karyotype if it were not the result of a singular chromosomal structural event? Surely, from what we now know of human cytogenetics, the frequency of fusion is uncommon enough to hypothesize that it occurred in several chimpanzees at the same time, animals that then got together for further breeding and speciation. The concept of ‘reticulate speciation’ of primates in general and humans specifically has been beautifully explored in the review by Arnold & Meyer (2006). It would appear to be a prominent process in mammals and specifically in primate species and the study by Osada & Wu (2005) supports this notion.
Mammalian speciation is complex, as is well rehearsed in the contribution by Eichler & Sankoff (2003). But this does not diminish the importance of the singular fusion process, one that might rightly be considered to be a saltational event and one that must necessarily have led to an extreme demographic ‘bottleneck' that resulted a very small number of founding proto-humans. When, in the hominid ancestry, this might have occurred will be the focus of much future discussion and speculation, as molecular biological studies tend to focus on the results of the isolation and on subsequent mutational events (by mtDNA-, electrophoresis-, gene- studies). Goodman (1999) stated eloquently from his studies and review of primate taxonomic speculations that “… the results of molecular studies of primate phylogeny (…) challenge the traditional anthropocentric view that humans are very different from all other animals . ”
The considerations of a primary chromosomal rearrangement from chimpanzees to humans contrast sharply with the view that an ‘original Eve' did not exist, an aspect that was explored in some detail by Ayala (1995), although earlier (1975), he had stated that “ ..in saltational speciation, reproductive isolation was initiated by chromosomal rearrangements …”.
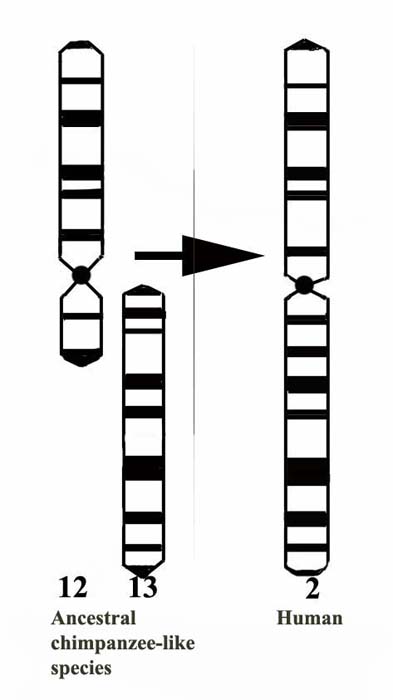 |
Figure 1. This represents the presumed ‘first step' in human evolution, the fusion (translocation) of the two chimpanzee submetacentric chromosomes ## 12 & 13. |
I will first next summarize some of the major evolutionary event affecting chromosomal organization in those mammalian species that have been studied in some detail, those that are most likely the results of major chromosomal rearrangements; they are largely comprised of fusions of acrocentric chromosomes (Robertson, 1916). The list is by no means limited to these few examples. Many other well-explored taxa ( e.g . Perissodactyla) follow a similar path. They are all presented by King in his comprehensive book (1993).
Apennine mice
Gropp (1969), in studying the captive population of the Tobacco mouse, Mus poschiavini , found a karyotype of 2n=26, while most European feral mice possess 2n=40 and which are composed of acrocentric chromosomes (Figure 3). This discovery followed by the subsequent backcrosses with ‘regular mice' led to a great variety of aneuploid offspring. This thus essentially prevented further gene flow to continue with the original population. These investigations have had a major impact for our understanding of the rapid chromosomal ‘speciation' process that has taken place in Alpine mice with their many different types of ‘Rb-chromosomes'.
Since that original discovery, mice from numerous other valleys of the Swiss/Italian Alps have been studied and a great variety of additional species, with different chromosome numbers due to various Robertsonian fusions of chromosomes, have been identified. They have also formed the basis of Capanna's extensive further research (Gropp et al., 1982, 1983; Capanna, 1982; Castiglia & Capanna, 1999). Most recently, additional examples of this fusion mechanism in mice have been discovered on the Mediterranean island of Madeira (Britton-Davidian et al., 2000), and others exist in Tunisia . Figure 2 shows a simplified drawing of this mechanism. As will be seen from this drawing, two principal modes – fission and fusion – are depicted. While the fusion concept is often witnessed, it even occurs in humans (see Shaffer & Lupski, 2000), the fission of metacentric chromosomes has been much more controversial, as it invariably implies the creation of new centromeres. The fate of centromeres and what drives their repositioning and complexity in offspring of hybrids has been admirably reviewed for mammals, but especially for macropods by O'Neill et al. (2004). It is safe to say now that no complete agreement has yet been reached as to the chromosomal structural or genetic causes of the events that lead to neo-centromeres, their duplication or repositioning as it may have occurred in lemurs for instance. It could be argued that the Apennine mice with their vastly different chromosomal arrays are merely ‘parapatric races' or ‘cytotypes' of Mus musculus domesticus, as over 100 different rearrangements have now been identified (Nachman et al., 1994). But, some and perhaps most investigators have denominated them as being good species. At the minimum, they represent examples that reduced gene flow between ancestral and new types of mice and the rearranged animals are thus, in my view, truly new species. Dumas & Britton-Davididan (2002) examined the chiasma frequency of Rb mice from Tunisia and found a significant decrease in the number of chiasmata in both sexes.
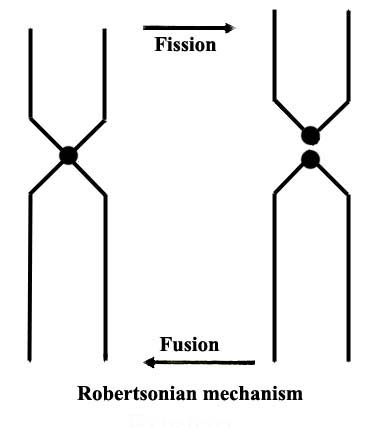 |
Figure 2. The essence of the acrocentric fusion/fission mechanism of Robertson (1916). |
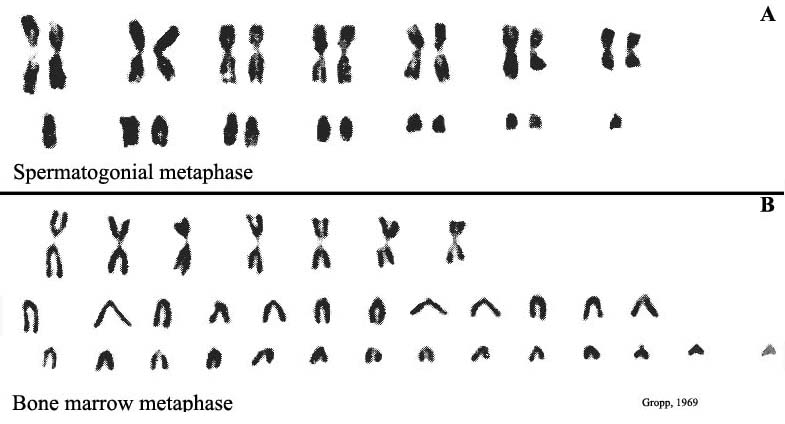 |
Figure 3. The original karyotypes of Mus poschiavini (from Gropp, 1969). |
Rupicaprini
This distinct tribe of animals extends from Europe (as chamois) to the New World (as Rocky Mountain goat), with many intermediate species ranging over Asia . Dolan (1963) has traced the evolution of these animals in a searching inquiry that has formed the basis for our interest in their chromosomal compositions. Figure 4 shows our findings of the continuous and gradual reduction of their chromosome numbers that we identified. We believe that they must have occurred through the process of fusion rearrangements, with resulting karyotypes from 2n=58 to 2n=42 and formation of new species.
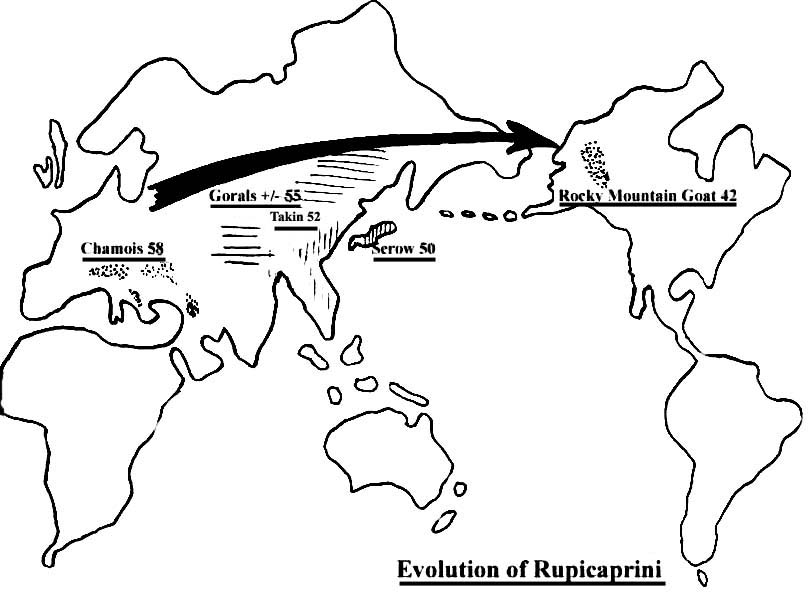 |
Figure 4. Evolution of Rupicaprini and their chromosome numbers. |
African gazelles
A very similar process of fusions probably occurred following the immigration of Gazellinae from Asia to Africa , with much speciation occurring subsequently on that new continent (Figures 5, 6). In this group of animals, an additional unusual event took place aside from the autosomal fusions with their reduction of chromosome numbers. That additional feature is the fusion of an autosome with sex chromosomes, thus making the variation even greater (Vassart et al., 1995, among others).
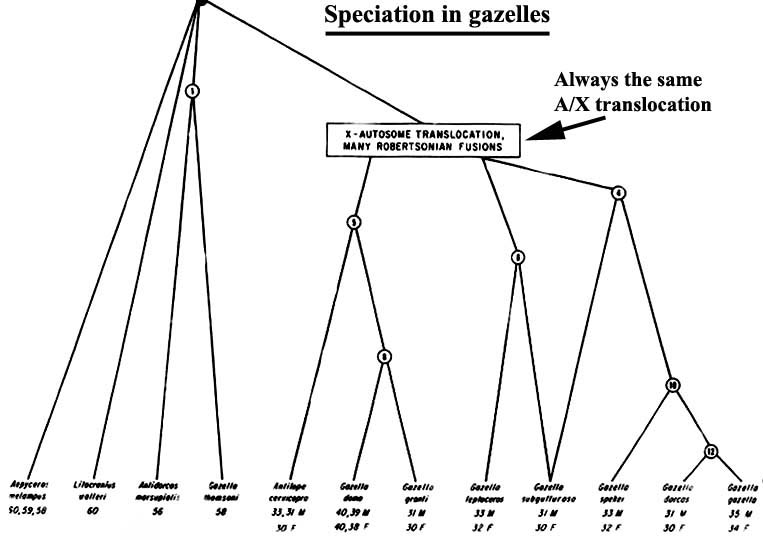 |
Figure 5. Speciation of all African gazelles, with same X/autosome fusion occurring in the right group, hence males and females have different numbers of chromosomes. Many subsequent fusions have also occurred. |
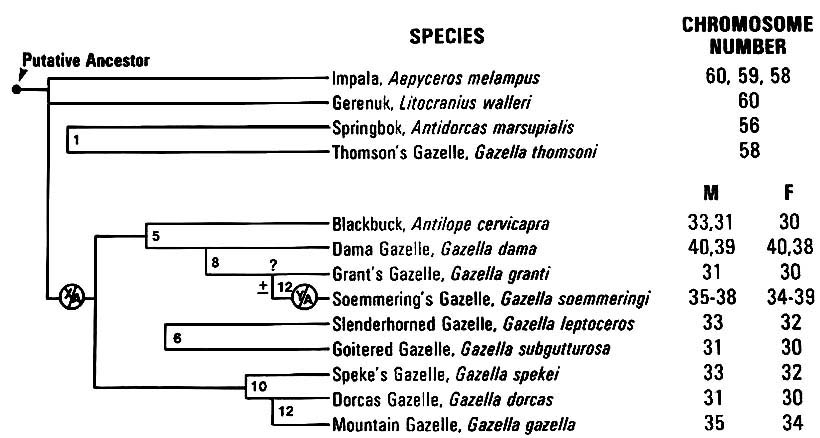 |
Figure 6. Similar arrangements as Figure 5. |
Mammalian Invasion into South America
When North and South America were joined around 2 MYA, numerous large animals invaded into South America through the developing land bridge, and some of them speciated after arriving on this new continent (Simpson, 1980). Because of this relatively recent event and judging from the study of their chromosomes, again chromosomal fusions must have played a significant role in limiting gene flow from the ancestral stock to the new species. Thus, several new feline species (ocelot, oncilla, marguay, Geoffrey's, Kodkod) evolved chromosome numbers of 2n=36 (all others Felidae have 2n=38). Ursidae and Artiodactyla followed a similar path. Thus, all bears studied have had chromosome numbers of 2n=74, only the South American spectacled bear has 2n=52. The North American deer have 2n=70, the brocket deer of South America that derived from it has 2n=50. And so are the peccaries rearranged, from 2n=30 in the collared peccary, via 2n=26 in the white-lipped peccary, to 2n=20 in the Paraguayan peccary, the ‘Tagua'. Further, all dogs have 2n=78, but the South American bush dog has 2n=74. Thus on two different continents and in several different mammalian taxa, recent colonization and speciation seem to have been accompanied by chromosomal fusion events.
What about Man?
It is now no longer seriously questioned that man evolved from chimpanzee-like ancestors with whom we share most of our genetic makeup. The details of these evolutionary relationships are often debated and they have been widely published in the scientific and lay presses. But, how did this process commence, the process that separated us from our ancestral species, Pan troglodytes , whose chromosome number remains at 2n=48?
I suggest that one ancestral hominoid experienced one major chromosomal fusion event, one that took place between chimpanzee chromosomes #12 and #13, thereby producing the new human chromosome #2. This initial “ Eve ” (or Adam) thus possessed 2n=47 chromosomes and then formed a ‘nuclear family'. During gametogenesis of such a heterozygous fusion individual, the segregation of chromosomes leads to individuals with 48, 47, and 46 chromosomes. Gene flow from ancestral hominoids would have continued. It is hypothesized that it may have occurred for an unknown length of time in this new ‘family'. Thus, the incorporation of much additional genetic material from hominoid relatives was possible. Nevertheless (and presupposing absence of major deleterious genetic abnormalities), numerous individuals would ultimately arise that possess the new 2n=46 karyotype. This event would then add some additional degree of reproductive barrier to chimpanzee ancestors, as many aneuploids would likely have been produced from continued hybridization with stock hominoids. This “Neo-Homo” then would have had a chance to evolve from this putative nuclear family, especially if one additionally assumes that some physical separation (? as perhaps movement into the savannah) and/or behavioral change may have occurred simultaneously. Patterson et al. (2006) have accrued much new evidence that has suggested prolonged hybridization of chimpanzees with early ‘hominoids’ and showed that much less genetic change has been taking place on the X-chromosome, perhaps as following “Haldane’s Rule”.
The effect on chromosomal meiotic segregation by both, a reciprocal translocation as well as consequences of Robertsonian fusions, is complex and has been explored by King (1993, pp. 77-84). In the simplest case, 50% aneuploid and 50% euploid gametes may arise, but this much depends on numerous factors that were extensively detailed by King. It is essentially impossible to predict these without knowing the defined breakpoints and a comprehensive study of meiosis, studies that have been done in a few species. Safe to say though, euploidy can be one sequel and King is explicit in stating (p. 122) that “... there is no longer any room for debate as to whether profoundly negatively heterotic chromosomal rearrangements can reach fixation in derived populations …”.
Given this putative scenario, additional subsequent chromosomal events (inversions etc.), as well as many new mutations could then take place and become fixed in NeoHomo , as gene flow became now significantly reduced until it completely stopped. One such chromosomal rearrangement, a pericentric inversion of chromosome 18, was well-defined by McConkey (1997); this inversion is not too dissimilar from the pericentric inversion of #2 in orang utans and is one that has not presented a barrier to ‘hybridization' (Figure 7) in orang utans.
The large number of mutations that have accumulated in humans since the separation from the lineage leading to chimpanzee is now legion. Lewontin's (1974) review of the application of gel electrophoresis has shown that an enormous genetic variation exists in humans, and in other mammalian populations as well. One of the perhaps more significant mutations may be the single copy gene CMAH which codes for the enzyme responsible for modifying the common cell surface sugar sialic acid from the precursor N-acetylneuraminic acid to the non-human form N-glycolylneuraminic acid that is common in other mammals, as aptly summarized by Varki (2001). But, numerous mutations are likely to have occurred in the chimpanzees after the split as well. In fact, when we now compare the ‘chimpanzee' with the human genome, we do not know precisely whether these chimpanzee genomes are at all comparable to what had existed when we ‘started out'. Indeed, the recent description by McBrearty & Jablonski (2005) of the first fossil chimpanzee teeth indicates that there is a possibility that some of the chimpanzee ‘forms' (? species), as well as many fossil ‘intermediate human species' have become extinct, because the extant chimpanzees from different regions in Africa are even now moderately dissimilar from one another. Ely et al. (2005) have recently studied some of the mtDNA variation in extant subspecies of chimpanzees and found that, despite differences, there was no problem with cross-breeding these varieties. In addition, the mtDNA studies published by Gagneux et al. (1999) show more mtDNA variation in chimpanzee and bonobo clades than seen in the entire human species. Shipman (2003) reviewed the African heritage of mtDNA and concluded from these then authoritative studies, that the original ‘Eve” (mtDNA having a maternal lineage and relatively constant known mutation rate) originated about 170,000 years ago.
Numerous studies of large human populations have shown that minor chromosomal variants (as identified by banding studies) exist; Wyandt & Tonk (2004) estimated them to be as common as occurring in 1% of the human population. In a separate chapter, Patil & Wyandt (2004) presented a review of heteromorphisms and aneuploidies seen in couples with infertility and, of course, they are even more common in neoplasms. The various papers cited are not all in agreement that infertility is a regular sequel of aneuploid parents. Robertsonian translocations (fusions) of human # 13 is reasonably common in ‘normal humans' and it appears to be a non-random event that has caused them; other acrocentric chromosomes are also often involved, such as with #14 and #21, and #14 with #21 being the commonest (Nagel & Hoehn, 1971). Interestingly, monocentrics more commonly evolve with fusion of homolog chromosomes, while dicentrics occur with heterologous fusion of chromosomes (p. 204 in Wyandt & Tong, 2004).
Although the Robertsonian model of speciation envisages also that fissions may occur, only very few examples (8) of fission have been reported in the vast number of human cytogenetic studies. Most of these were of deformed fetuses, only a few occurred in families (Bogart et al., 1995; Perry, 2004). The problem of neocentromeres production that is implicit with the fission event has been explored by Warburton (2004). Inversions in several chromosomes (## 1, 2, 3, 9, 10, 16, 18, 19, Y) have also been observed in the human population and they are generally not associated with phenotypic abnormalities (Verma et al., 1978)
Implicit in these considerations is the idea that extremely small populations form the nucleus of a new species. This, however, runs counter to the intuitive notion of ‘ inbreeding depression' , a concept that is especially pervasive in the zoological community. It should be noted, however, that the terms ‘inbreeding, population size, etc.' have been used to apply to many different concepts, as is well discussed by Templeton & Read (1994) in the considerations of their successful management of Speke's gazelles at St. Louis Zoo. A founding population of 1.3 animals was there successfully reared following a defined protocol, one that was chosen to avoid drift. Over 100 Speke's gazelles have since been born, but new stock has also recently been added. The model is not completely useful as the varied maintenance conditions at different zoos and other factors tend to muddy the analysis. Nevertheless, the captive population has not become so inbred as to come near extinction from ‘inbreeding depression'. Numerous other examples of a very small founder population can be cited that resulted in a huge expansion of animals, one that is not too dissimilar from what must have occurred with the “bottleneck” of our human ancestry. Most other animals cited below were randomly bred and they still succeeded with their expansion. The notion of an initially small population as starting point and the future expansion have been beautifully illustrated by Ernst Mayr in the next Figure
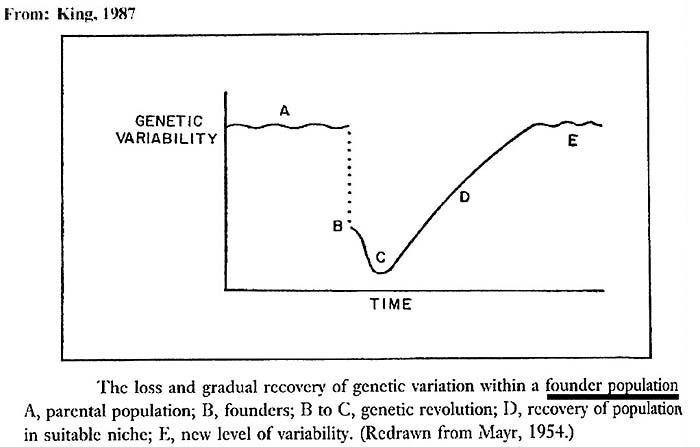 |
Figure 7. Mayr's concept of the event that follow a founder event. |
‘Genetically disease-free' individuals must be rare, however. It is generally assumed, for instance, that each of us possesses 4-5 lethal mutations that necessarily occur in a heterozygous state. Perhaps this factor alone is the major reason why speciation is so rarely witnessed in mammals or why it may be such an uncommon event in general even though chromosomal rearrangements may be common. To succeed with the event of chromosomal evolution as here presented, the ‘inbreeding' necessary would often be fatal for any arising group of animals, mostly because of the prevalence of extant lethal mutations. And that alone may be the reason why it is so rarely witnessed. An alternate view was presented by Spielman et al. (2004), however, in their study that employs a meta-analysis of heterozygosity loci in endangered species vs . those of non-endangered species. It showed that the former have a much lower rate of heterozygosity. They inferred from these data that the loss of heterozygosity greatly impacts threatened species.
Thus, another possible reason why speciation is so rare could be that each new species starts out as an extremely “low heterozygosity” species and that nascent species are by definition highly endangered and probably die out more often than they survive.
This is countered on the other hand by numerous examples of such ‘bottlenecks' that have existed in the ancestry of sometimes very large populations of mammals, some of which are summarized next. These observations argue strongly against the correctness of the notion that inbreeding depression befalls all species. All the modeling and computer-generated predictions of doom thus may not necessarily hold true for all small founder-derived populations. Moreover, in some instances ‘outbreeding depression' (mixing with chromosomally different populations of said-to-good species [Dik dik]) has been the real problem of the crashes witnessed in some zoological parks.
1) Golden hamster - all millions extant now come from the offspring of one pregnant female caught in Aleppo .
2) Nilgai - 4.8 released in Texas in 1941; now 9,000+, annual harvest 1,000+.
3) Arabian oryx – 5 founders: 220+ born since. Reintroduced.
4) Slender-horned gazelle – 3 founders: now 100+ living.
5) Persian gazelle - 3 founders: 600 + born.
6) Przewalski's horse - 7.6 founders: thousands born, 248 animals reintroduced to Mongolia. ( San Diego alone has bred 125 animals). The mortality in our stock has been largely due to management problems (lack of avoidance of coccidioidomycosis).
7) Père David's deer - 3 founders from China : 700+ living.
8) Mhorr gazelles - 1.5 founders rescued by Spanish military man, extinct in wild, more than 230+ bred.
9) Chinchilla -11 animals imported in 1923 by a mining engineer from LA – millions were bred since.
10) Muskrat - in 1905 very few animals escaped in Europe and are now a great pest there, with three conferences held how to eliminate them from the countryside.
11) Speke's gazelle . 1.3 animals to St. Louis , over 100 bred before new stock was added, no known genetic problems.
Birds:
11) Crested ibis ( China , Korea ) - the Internatl. Crane Fndtn. rescued 7 animals, bred over400, released 300 into habitat, doing well. Was thought to be extinct.
12) Nene goose ( Hawaii ) - rescued by one man into Slimbridge (UK).
13) California condor - 22 ancestors: 243 bred, some released (an anomaly of cartilage formation exists, however)
The following animals started with these low numbers and are now great pests in Australia (and the evidence shows that the rabbit has adapted to the introduced myxomatosis virus – as has the virus):
| Bennett's wallaby – 6 | Sambar deer - 2 |
| Hares (Rabbit) – 6 | Rusa deer - 8 |
| Chamois – 8 | Sika deer – 6 |
Summary of some major animal population that successfully weathered a major bottleneck.
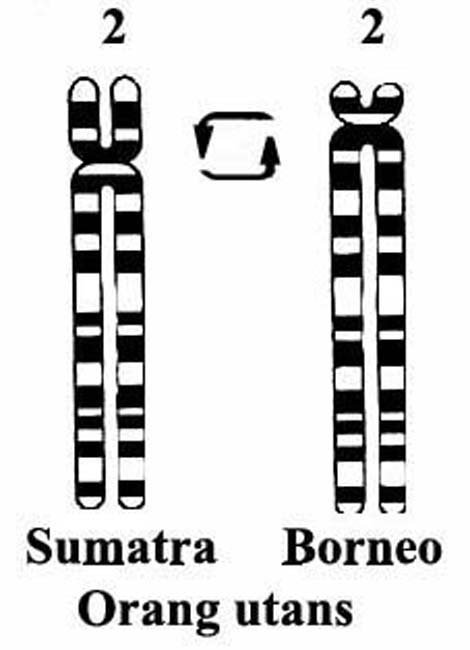 |
Figure 8. Orangutan pericentric inversion of chromosome #2. |
How frequent are chromosomal rearrangements in human studies?
A survey of 14,069 newborn infants' chromosomes by Hamerton et al. (1975) indicated that 1:150 - 1:200 live-born babies have major chromosomal abnormalities, including Robertsonian and reciprocal translocations. Some of these are lethal, but rearrangements of chromosomes in the general human population are also significant and might possibly relate to the future evolutionary development in humans. Shaffer & Lupski (2000) have summarized the numbers of major chromosomal variations in human populations; these are shown next.
Frequency of human chromosomal rearrangements
| Type of rearrangement | Frequency in general population |
| Reciprocal translocations | 1 in 625 |
| Robertsonian translocations | 1 in 1000 |
| Marker chromosomes | 1 in 2000 |
| Interstitial deletions | 1 in 4000 (at least) |
| Interstitial duplications | 1 in 4000 (at least) |
| Terminal deletions | 1 in 5000 (at least) |
Incidence of human chromosomal rearrangements during female meiosis (Shaffer & Lupski, 2000).
How could one prove that this concept of chromosomal speciation applies to mammals?
Aside from the numerous models of speciation developed in Drosophila, it would be nice to witness the event in mammals directly. Mice would probably be the most suitable species because of their short generation time and their relatively frequently occurring Robertsonian fusions. Thus, Harris et al. (1986) witnessed a Robertsonian translocation in their mouse colony, ascertained a nondisjunction rate in the male of 10% and estimated that 12-16% of trisomic or monosomic zygotes occurred in the heterozygous females; in addition there was a significant excess of heterozygotes in phenotypically normal offspring. Breeding such heterozygotes long enough should eventually produce homozygotes for the fusion chromosome and then the backcross disturbance or genetic ‘isolation' to ancestral mice could be studied. Exposure of this new homozygote stock to mild mutagens such as ENU (Guénet, 2004) just might enhance the possibility of creating new ‘strains' and, if one does this long enough, perhaps a new species actually evolves – who knows? Under current study is the outcome of the ‘tension zones’ that exist in chromosomally divergent ‘species/races’ of the common shrew (Sorex araneus) see: Hatfield et al., 1992, others.
References:
Arnold, M.L. and Meyer, A.: Natural hybridization in primates: One evolutionary mechanism. Zoology 109:261-276, 2006.
Ayala, F.J.: Genetic differentiation during the speciation process. Evolut. Biol. 8:1-78, 1975.
Ayala, F.J.: The myth of Eve: Molecular biology and human origins. Science 270:1930-1936, 1995.
Ayala, F.J. and Coluzzi, M.: Chromosome speciation: Humans, Drosophila, and mosquitoes, pp.46-68 in: Hey, J., Fitch, W.M. and Ayala, F.J.: Systematics and the Origin of Species. On Ernst Mayr's 100 th Anniversary. National Academy of Sciences , USA , Washington , D.C. 2005.
Barton, N.H.: The role of hybridization in evolution. Mol. Ecol. 10:551-568, 2001.
Baker, R.J. and Bradley, R.D.: Speciation in mammals and the genetic species concept. J. Mammal. 87:643-662, 2006.
Benirschke, K. and Miller, C.J.: Anatomical and functional differences in the placenta of primates. Biol. Reprod. 26:29-53, 1982.
Bogart, M.H., Fujita, N., Serles, L. and Hsia, Y.E.: Prenatal diagnosis of a stable de novo centric fission: a case report. Amer. J. Med. Genet. 59:36-37, 1995.
Brinn, M.N.A., Porto , J.I.R. and Feldberg, E.: Karyological evidence for interspecific hybridization between Cichla monoculus and C. temensis (Perciformes, Cichlidae) in the Amazon. Hereditas 141:252-257, 2004.
Britton-Davidian, J., Catalan, J., da Graca Ramalhinho, M., Ganem, G., Auffray, J.C., Capela, R., Biscoito, M., Searle, J.B. and da Luz Mathias, M.: Rapid chromosomal evolution in island mice. Nature 403:158, 2000.
Capanna, E.: Robertsonian numerical variation in animal speciation.: Mus musculus , an emblematic model. Pp. 155-174, in Mechanisms of Speciation, C. Barigozzi, ed. A. Liss, N.Y. 1982.
Carter, A.M. and Enders, A.C.: Comparative aspects of trophoblast development and placentation. Reprod. Biol. Endocrinol. 5:46, 2004. (Review).
Castiglia, R. and Capanna, E.: Whole-arm translocation (WART) in a feral population of mice. Chromosome Res. 7:493-495, 1999.
Chiarelli, A.B. and Capanna, E.: Cytotaxonomy and Vertebrate Evolution. Academic Press, London and New York, 1973.
Coyle, J.A. and Orr, H.A.: Speciation. Sinauer Associates, 2004.
Crow, T.J.: The Speciation of Modern Homo sapiens. Oxford University Press, Oxford, UK, 2002.
Darwin, C.: The Origin of Species and the Descent of Man. 1859. Reprinted by: The Modern Library NY.
Dolan, J.M.: Beitrag zur systematischen Gliederung des Tribus Rupicaprini Simpson, 1945. Z. zool. Syst. Evolutionsforsch ung 1, 3-4, 311-407, 1963. Akademische Verlagsgesellschaft, Frankfurt/Main.
Dumas, D. and Britton-Davidian, J.: Chromosomal rearrangements and evolution of recombination: comparison of chiasma distribution patterns in standard and Robertsonian populations of the house mouse. Genetics 162:1355-1366, 2002.
Eichler, E.E. and Sankoff, D.: Structural dynamics of eukaryotic chromosome evolution. Science 301:793-797, 2003.
Eldredge, N. and Gould, S.J.: Punctuated equilibria: An alternative to phyletic gradualism. Pp. 82-115 in, Models of Paleobiology, T.J.M. Schopf, ed. Freeman, Cooper and Co., 1972.
Ely, J.J., Dye, B., Frels, W.I., Fritz, J., Gagneux, P., Khun, H.H., Switzer, W.M. and Lee, D.R.: Subspecies composition and founder contribution of the captive U.S. chimpanzee (Pan troglodytes) population. Amer. J. Primatol. 67:223-241, 2005.
Erlich, H.A., Bergström, Stoneking, M. and Gyllensten, U.: HLA sequence polymorphism and the origin of humans. Science 274:1552-1554, 1996 (with Ayala's reply).
Gagneux, P., Wills, C., Gerloff, U., Tautz, D., Morin, P.A., Boesch, C., Fruth, B., Hohmann, G., Ryder, O.A. and Woodruff, D.: Mitochondrial sequences show diverse evolutionary histories of African hominoids. Proc. Natl. Acad. Sci. USA 96:5077-5082, 1999.
Gelder, R. van: Mammalian Hybrids and Generic Limits. Novitates. American Museum . No. 2635, pp. 1-25, October 12, 1977 .
Genner, M.J. and Turner, G.F.: The mbuna cichlids of Lake Malawi : a model for rapid speciation and adaptive radiation. Fish and Fisheries 6:1-34, 2005.
Goodman, M.: Molecular evolution '99. The genomic record of humankind's evolutionary roots. Amer. J. Hum. Genet. 64:31-39, 1999.
Gould, S.J.: The Structure of Evolutionary Theory. The Belknap Press of Harvard University Press, Cambridge, Massachusetts, 2002.
Gropp, A.: Cytologic mechanisms of karyotype evolution in insectivores. Pp. 247-266, in: Comparative Mammalian Cytogenetics, K. Benirschke, ed. Springer-Verlag, NY, 1969.
Gropp, A., Winking, H., Redi , C.A. , Capanna, E., Britton-Davidian, J. and Noack, G.: Robertsonian karyotype variation in wild house mice from Rhaeto-Lombardia. Cytogenet. Cell Genet. 34:67-77, 1982.
Gropp, A., Winking, H., Herbst, E.W. and Claussen, C.P.: Murine trisomy: Developmental profiles of the embryo, and isolation of trisomic cellular systems. J. Exp. Zool. 228:253-269, 1983.
Grosser, O.: Frühentwicklung, Eihautbildung und Placentation des Menschen und der Säugetiere. J.F. Bergmann, München, 1927.
Groves, C.: Primate Taxonomy. Smithsonian Institution Press, Washington , DC , 2001.
Guénet, J.L.: Chemical mutagenesis of the mouse genome: An overview. Genetica 122:9-24, 2004.
Hamerton, J.L., Canning, N., Ray, M. and Smith, S.: A cytogenetic survey of 14,069 newborn infants. I. Incidence of chromosome abnormalities. Clin. Genet. 8:223-243, 1975.
Harris, M.J., Wallace , M.E. and Evans, E.P.: Aneuploidy in the embryonic progeny of females heterozygous for the Robertsonian chromosome (9.12) in genetically wild Peru-Coppock mice (Mus musculus). J. Reprod. Fertil. 76:193-203, 1986.
Hatfield, T., Barton, N. and Searle, J.B.: A model of a hybrid zone between two chromosomal races of the common shrew (Sorex araneus). Evolution 46:1129-1145, 1992.
King, M.: Species Evolution. The Role of Chromosome Change. Cambridge University Press, Cambridge, UK, 1993.
Kirschner, M.W. and Gerhart, J.C.: The Plausibility of Life. (Resolving Darwin 's Dilemma). Yale University Press, New Haven and London, 2005.
Lande, R., Seehausen, O. and van Alphen, J.J.M.: Mechanism of rapid sympatric speciation by sex reversal and sexual selection in cichlid fish. Genetica 112-113:435-443, 2001.
Lewontin, R.C.: The Genetic Basis of Evolutionary Change. Columbia University Press, New York, 1974.
Mayr, E.: Animal Species and Evolution. Belknap Press of Harvard University Press, Cambridge, Massachusetts, 1963.
McBrearty, S. and Jablonski, N.G.: First fossil chimpanzee. Nature 437:105-108, 2005.
McConkey, E.H.: The origin of human chromosome 18 from a human/ape ancestor. Cytogenet. Cell Genet. 76:189-191, 1997.
Mossman, H.W.: Vertebrate Fetal Membranes. MacMillan, Houndmills, 1987.
Nachman, M.W., Boyer, S.N., Searle, J.B. and Aquadro, C.F.: Mitochondrial DNA variation and the evolution of Robertsonian chromosomal races of house mice, Mus domesticus . Genetics 136:1105-1120, 1994.
Nagel, M. and Hoehn, H.: On the non-random involvement of D-group chromosomes in centric fusion translocations in man. Humangenetik 11:351-354, 1971.
O'Neill, R.J., Eldridge, M.D.B. and Metcalfe, C.J.: Centromere dynamics and chromosome evolution in marsupials. J. Hered. 95:375-381, 2004.
Osada, N. and Wu, C.I.: Inferring the mode of speciation from genomic data: a study of the great apes. Genetics 169:259-264, 2005.
Patil, S.R. and Wyandt, H.E.: Heteromorphisms in clinical populations. Chapter 4, pp. 47-62, in, Atlas of Human Chromosome Heteromorphisms, Wyandt, H.T and Tonk, V.S., eds., Kluwer Academic Publishers, Dordrecht, Boston, London, 2004.
Patterson, N., Richter, D.J., Gnerre, S., Lander, E.S. and Reich, D.: Genetic evidence for complex speciation of human and chimpanzees. Nature 441:1103-1108, 2006.
Pennisi, E: Epigenetics. Behind the scenes of gene expression. Science 203:1064-1067, 2001.
Perry, J., Slater, H.R. and Choo, K.H.: Centric fission – simple and complex mechanisms. Chrom. Res. 12:627-640, 2004.
Robertson, W.: Chromosome studies. I. Taxonomic relationships shown in the chromosomes of Tettigidae and Acrididae . V-shaped chromosomes and their significance in Acrididae, Locustidae, and Grillidae : chromosomes and variation. J. Morphol. 27:179. 1916.
Ruse , M.: Darwin and Design. Does Evolution have a purpose? Harvard University Press, Cambridge , Massachusetts , 2003.
Ruse , M.: The Evolution-Creation Struggle. Harvard University Press, Cambridge , Massachusetts , 2005.
Ryder, O.A. and Chemnick, L.G.: Chromosomal and mitochondrial DNA variation in orang utans. J. Hered. 84:405-409, 1993.
Seehausen, O., van Alphen, J.J.M. and Witte, F.: Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science 277:1808-1811, 1997.
Shaffer, L.G. and Lupski, J.R.: Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu. Rev. Genet. 34:297-329, 2000.
Shipman, P.: We are all Africans. Amer. Scientist 91:496-499, 12003.
Simpson, G.G.: Splendid Isolation. The curious History of South American Mammals. Yale University Press, New Haven , Connecticut , 1980.
Spielman, D., Brook, B.W. and Frankham, R.: Most species are not driven to extinction before genetic factors impact them. Proceed. Natl. Acad. Sci. USA 101:15261-15264.
Tattersall, I. : The case for saltational events in human evolution. Pp. 49-59, in: The Speciation of Modern Homo sapiens . T.J. Crow ed., Oxford University Press, New York, 2002-2003. and Proc. Brit. Academy 106:49-49, 2002.
Templeton, A.R.: Gene lineages and human evolution. Science 272:1363-1364, 1996 (with Ayala's reply).
Templeton, A.R. and Read, B.: Inbreeding: One word, several meanings, much confusion. Pp. 91-105 in, Conservation Genetics, V. Loeschcke, J. Tomiuk and S.K. Jain, eds. Birkhäuser Verlag, Basel, Switzerland, 1994.
Varki, A.: Loss of N-Glycolylneuraminic acid in Humans: Mechanisms, consequences, and implication for hominid evolution. Yearb. Phys. Anthropol. 44:54-69, 2001.
Vassart, M., Seguela, A. and Hayes, H.: Chromosomal evolution in gazelles. J. Hered. 86:216-217, 1995.
Verma, R.S., Dosik, H. and Lubs, H.A.: Size and pericentric inversion heteromorphisms of secondary constriction regions (h) of chromosomes 1, 9 and 16 as detected by CBG technique in Caucasians: classification, frequencies and incidence. Amer. J. Med. Genet. 2:331-339, 1978.
Warburton, P.: Chromosomal dynamics of human neocentromeres formation. Chrom. Res. 12:617-626, 2004.
Weiner, J.: Evolution in action. Natural History, Nov. 2005, pp 47-51. 2005.
White, M.J.D.: Modes of Speciation. W.H. Freman and Co., San Francisco , 1978.
Wyandt, H.E. and Tonk, V.S.: Normal population studies. Chapter 3, pp. 33-46, in, Atlas of Human Chromosome Heteromorphisms, Wyandt, H.T and Tonk, V.S., eds., Kluwer Academic Publishers, Dordrecht, Boston, London, 2004.